The intricate process of duplicating genetic information, referred to as DNA replication, lies at the heart of the transmission of life from one cell to another and from one organism to the next. This happens by not just copying the genetic information; a well-orchestrated sequence of molecular events must also happen at the right time.
Scientists working with Prof. Maria-Elena Torres-Padilla from Helmholtz Munich have recently uncovered a fascinating aspect of this process known as “replication timing” (RT) and how special this is when life commences. The new results are now published in Nature.
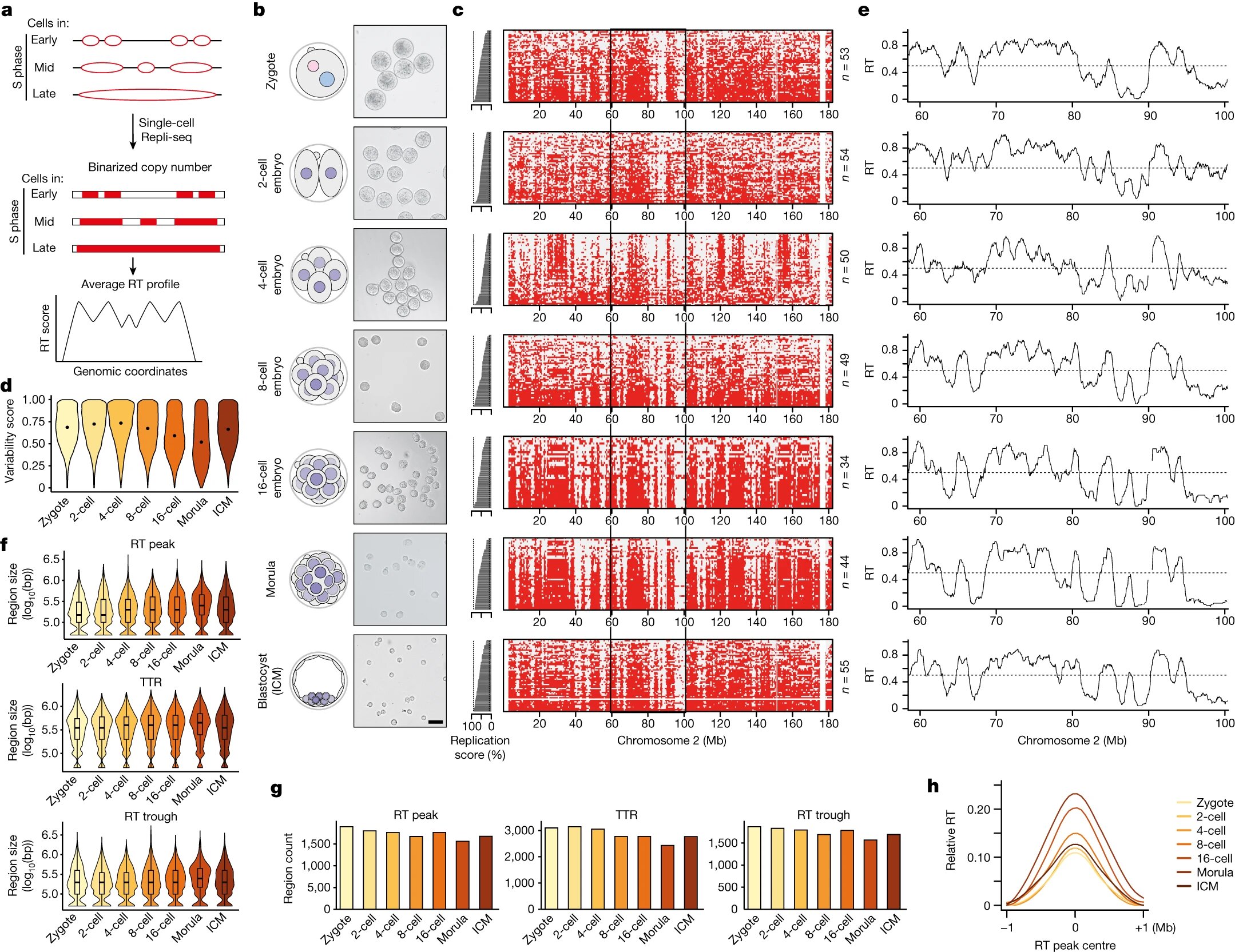
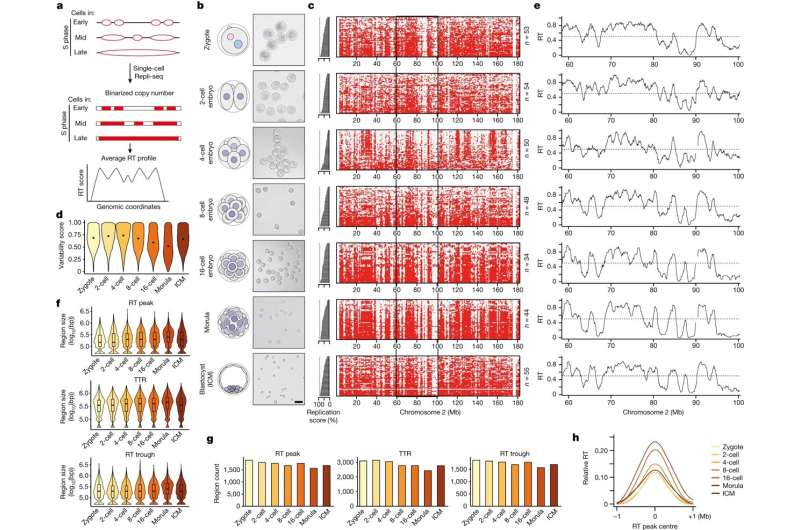
The process of DNA replication timing (RT) refers to the specific moments when different regions of our genetic code are duplicated. Researchers from the Institute for Epigenetics and Stem Cells at Helmholtz Munich have implemented a technique called “Repli-seq” to delve into the intimate relationship between RT and the adaptability of cells, the cellular plasticity.
Intriguingly, they also uncovered a new relationship between RT and how the genes fold into three-dimensional structures inside the cell nucleus.
Starting with the earliest stage of an embryo, the zygote—the very beginning of an organism’s life—researchers have created a map of RT from this single-cell stage to the stage at which the embryo implants in the mother’s womb, called a blastocyst. The unexpected discovery is that the RT in the single-celled embryo is not very ordered, suggesting that genome duplications are very flexible in these early cells.
However, after the 4-cell stage, the RT becomes more defined. A gradual process is happening, mirroring the gradual acquisition of modifications to the DNA and associated proteins, the so-called chromatin marks, that indicate the genes’ activity and importance in the cell’s functions.
Maria-Elena Torres-Padilla, corresponding author of the study, explains further, “This is remarkable, as this tells us that these early embryo cells have a very ‘plastic’ genome duplication program. Because these early cells are totipotent, they can create every single cell in our bodies. We think that what we discovered in this study is one of the reasons why these cells are so remarkably capable of generating all the body.”
The new findings about DNA replication can be a tool to reprogram cells. Dr. Tsunetoshi Nakatani, the first author of the study, adds, “We can envision changing the cell identity by changing its RT program into a more flexible one.”
The results further show that RNA polymerase, commonly known as the enzyme responsible for reading the genetic code and transcribing it into RNA, contributes to determining the exact RT program, providing some cues as to how to be able to manipulate such a program in the future.
The research team has discovered that the three-dimensional structure of the genome takes shape first, and the RT program is established consequently. This is an exciting finding, as it posits that how our genome accommodates into the cell nucleus’s three-dimensional space influences the RT program’s flexibility.
In conclusion, DNA replication timing is a fascinating piece of the puzzle in the grand narrative of life. It demonstrates how the precision of genetic replication is intimately tied to the capacity of the cells from the early embryo to generate other cell types in our body. As researchers continue to explore these connections, we gain a deeper understanding of the very essence of life’s transmission, cell to cell, organism to organism, and of what makes a cell capable of generating a new body.
More information:
Maria-Elena Torres-Padilla, Emergence of replication timing during early mammalian development, Nature (2023). DOI: 10.1038/s41586-023-06872-1
Provided by
Helmholtz Association of German Research Centres
Citation:
Team discovers relationship between DNA replication timing and how genes fold into 3D structures inside cell nucleus (2023, December 20)
retrieved 20 December 2023
from https://phys.org/news/2023-12-team-relationship-dna-replication-genes.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.





