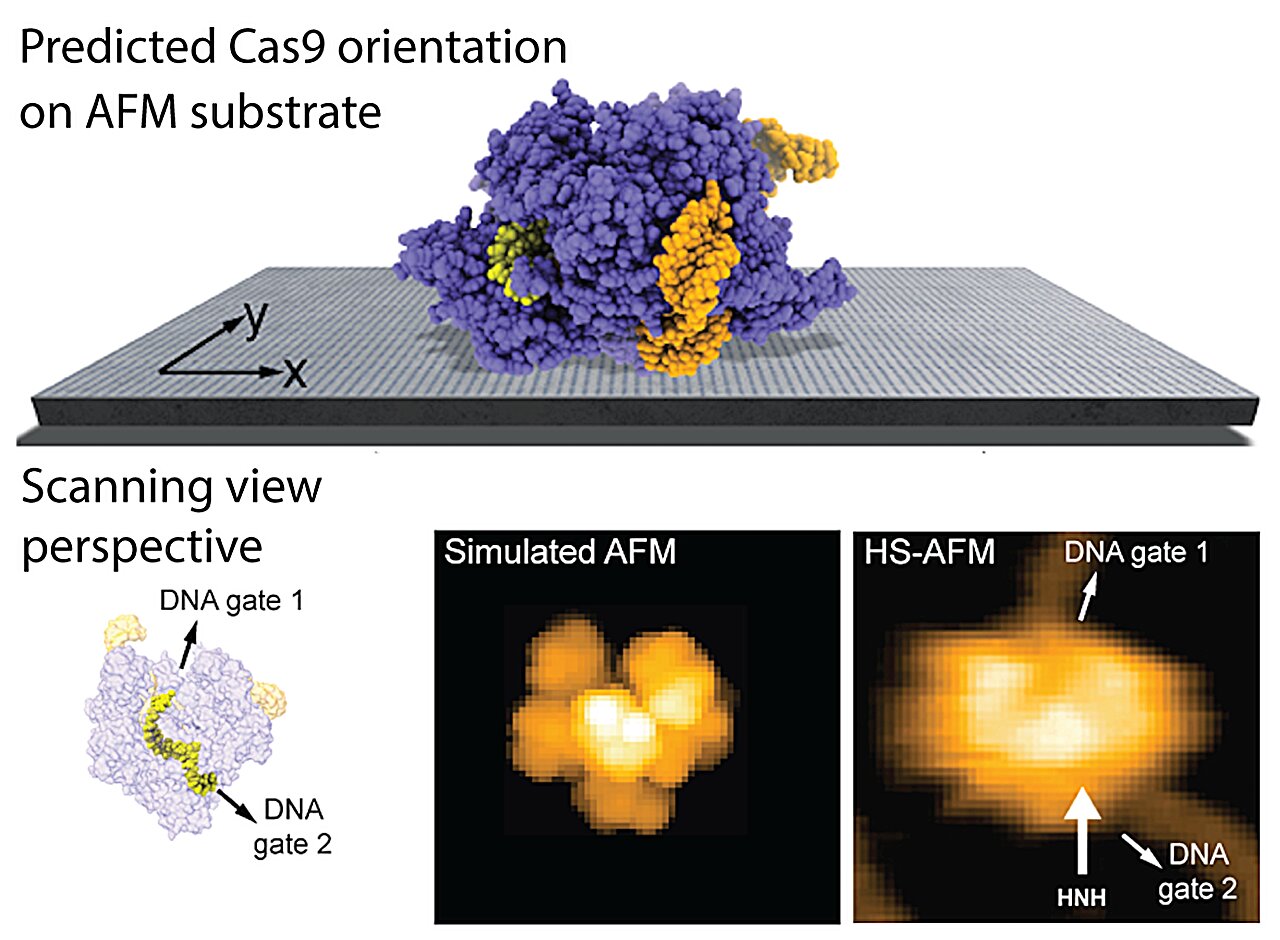
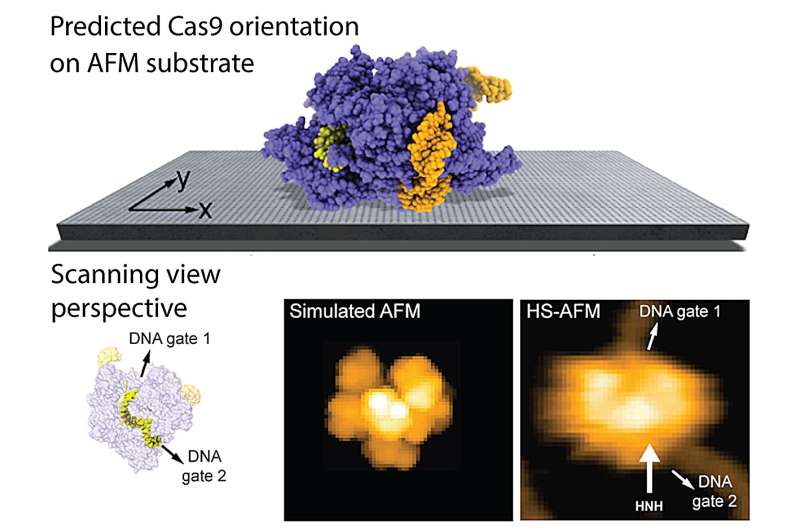
Researchers at Kanazawa University report in Frontiers in Molecular Biosciences a computational method to predict the placement of proteins on AFM substrates based on electrostatic interactions
The observation of biomolecular structures using atomic force microscopy (AFM) and the direct visualization of functional conformational dynamics in high-speed AFM (HS-AFM) experiments have significantly advanced the understanding of biological processes at the nanoscale.
In experiments, a biological sample is deposited on a supporting surface (AFM substrate) and is scanned by a probing tip to detect the molecular shape and its dynamical changes. The observation of protein dynamics under HS-AFM is a delicate balance between immobilizing the structure on the supporting surface while at the same time preventing too strong perturbations by immobilization.
The process of placing a biomolecular sample on the supporting surface and controlling its proper attachment is a challenge at the very start of every AFM observation. By the chemical composition of the buffer, interactions between the sample and substrate can be modified. Such surface modifications are often critical for successful AFM observations of protein structures and their functional motions. However, the molecular orientation of the sample is ‘a priori’ unknown, and due to limitations in the spatial resolution of images, difficult to infer from ‘a posteriori’ analysis.
Romain Amyot, Noriyuki Kodera, and Holger Flechsig from Kanazawa University have now developed a physical model to predict the placement of biomolecular structures on AFM substrates based on electrostatic interactions. The method considers the substrates commonly used in AFM experiments (mica, APTES-mica, lipid bilayers) and takes into account buffer conditions.
In computer simulations, a large number of possible molecular arrangements on the AFM substrate are sampled, and from evaluating the corresponding interaction energies, the most favorable placement is determined. Furthermore, the analysis allows predictions of the imaging stability under tip scanning.
The authors provide several applications of the new method and obtain remarkable agreement of model predictions with previous experimental HS-AFM imaging of proteins. The findings can explain, for example, why HS-AFM observations of the Cas9 endonuclease, a protein playing a key role in genetic engineering applications, can reliably visualize functional relative motions of target DNA and Cas9 and capture DNA cleavage events at the single molecule level.
Furthermore, as demonstrated for the ATP-powered chaperone machine ClpB, the model can explain how buffer conditions affect the stability of the sample-substrate complex and validate observations of previous HS-AFM experiments.
In summary, the new method allows to employ the enormous amount of available structural data for biomolecules to make predictions of the sample placement on AFM substrates even prior to an actual experiment, and it can also be applied for post-experimental analysis of AFM imaging data.
The developed method is implemented within the freely available BioAFMviewer software package, providing a convenient platform for applications by the broad BioAFM community.
More information:
Romain Amyot et al, Predicting the placement of biomolecular structures on AFM substrates based on electrostatic interactions, Frontiers in Molecular Biosciences (2023). DOI: 10.3389/fmolb.2023.1264161
Provided by
Kanazawa University
Citation:
Researchers predict protein placement on atomic force microscopy substrates (2023, December 13)
retrieved 13 December 2023
from https://phys.org/news/2023-12-protein-placement-atomic-microscopy-substrates.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.



